For BDS dataset, variables such as PARAMTYP and DTYPE may be required to be created when coming to complicated study. This post will introduce the difference among these two variables and how to develop them.
Introduction on PARAMTYP and DTYPE
Following table is from ADaM IG. You can see that PARAMTYP should be applied when you need to create a new parameter. Suppose that height and weight are collected in VS domain, but BMI (Body Mass Index) is not collected, what should you do if you need to develop a table which contains summary of BMI? In this case, we need to create a new record having PARAMCD equals to BMI (values of BMI should be derived based on HEIGHT and WEIGHT) for each subject, each visit. And for these new records, we have to populate PARAMTYP as DERIVED.
| Variable Name | Variable Label | Codelist Controlled Term/ | CDISC Notes |
| PARAMTYP | Parameter Type | (PARAMTYP) | Indicator of whether the parameter is derived as a function of one or more other parameters. |
| DTYPE | Derivation Type | (DTYPE) | Analysis value derivation method. DTYPE is used to denote, and is required to be populated, when the value of AVAL or AVALC has been imputed or derived differently than the other values within the parameter. DTYPE is required to be populated even if AVAL and AVALC are null on the derived record. |
As for DTYPE, here is a common senario. For example, HEMOGLOBIN is supposed to be collected at all visits from SCREENING, WEEK 12, WEEK 24, WEEK 48 to WEEK 72, but for one subject, HEMOGLOBIN is missing at visits WEEK 24 and WEEK 72. In this case, the missing value can be imputed as non-missing values from last visit. For these imputed values, we have to create a new record, give it a flag variable – DTYPE and populate DTYPE as LOCF (Last Observation Carried Forward).
Here is a summary of situations when DTYPE should be populated.
| 1 | A new row is added within a parameter with the analysis value populated based on other rows within the parameter, |
| 2 | A new row is added within a parameter with the analysis value populated based on a constant value or data from other subjects, |
| 3 | An analysis value (AVAL or AVALC) on an existing record is being replaced with a value based on a pre-specified algorithm. |
And here are the common DTYPE values.
| LOCF | Last Observation Carried Forward. |
| WOCF | Worst Observation Carried Forward. |
| AVERAGE | AVERAGE of values. |
Case 1: How to derive PARAMTYP
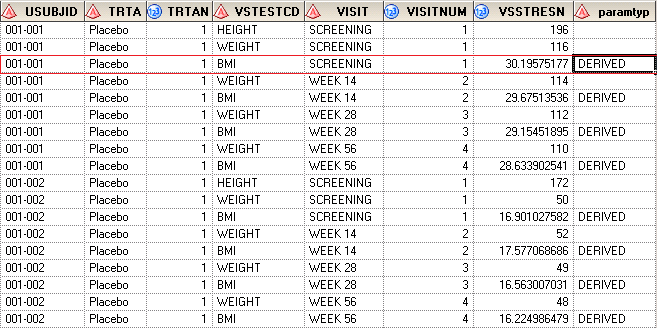
Suppose that we have WEIGHT and HEIGHT collected in CRF and need to derive BMI based on WEIGHT and HEIGHT. Here is the dummy data which contains in an excel file. The excel file named ADVS is stored in D:\ driver.
| USUBJID | TRTA | TRTAN | VSTESTCD | VSTEST | VISIT | VISITNUM | VSSTRESN | VSSTRESU |
| 001-001 | Placebo | 1 | HEIGHT | HEIGHT | SCREENING | 1 | 196 | cm |
| 001-001 | Placebo | 1 | WEIGHT | WEIGHT | SCREENING | 1 | 116 | kg |
| 001-001 | Placebo | 1 | WEIGHT | WEIGHT | WEEK 14 | 2 | 114 | kg |
| 001-001 | Placebo | 1 | WEIGHT | WEIGHT | WEEK 28 | 3 | 112 | kg |
| 001-001 | Placebo | 1 | WEIGHT | WEIGHT | WEEK 56 | 4 | 110 | kg |
| 001-002 | Placebo | 1 | HEIGHT | HEIGHT | SCREENING | 1 | 172 | cm |
| 001-002 | Placebo | 1 | WEIGHT | WEIGHT | SCREENING | 1 | 50 | kg |
| 001-002 | Placebo | 1 | WEIGHT | WEIGHT | WEEK 14 | 2 | 52 | kg |
| 001-002 | Placebo | 1 | WEIGHT | WEIGHT | WEEK 28 | 3 | 49 | kg |
| 001-002 | Placebo | 1 | WEIGHT | WEIGHT | WEEK 56 | 4 | 48 | kg |
Click here to hide/show code
proc import datafile=”D:\ADVS.xlsx”
out=ADVS
dbms=xlsx
replace;
sheet=”Sheet1″;
startrow=2;
getnames=Yes;
run;
Derive BMI based on (Screening) HEIGHT and WEIGHT
Here is again to demonstrate power of RETAIN statement and By statement. We can populate n as 1 for HEIGHT record and 2 for other records in order to make HEIGHT observation as the first record for each subject.
Click here to hide/show code
data advs;
set advs;
if vstestcd = “HEIGHT” then n = 1;
else n = 2;
run;
proc sort data=advs; by usubjid n; run;
data advs1(drop=height n);
length paramtyp $20.;
retain height;
set advs;
by usubjid n;
if first.usubjid and n = 1 then height = vsstresn;
if vstestcd = “WEIGHT”;
paramtyp = “DERIVED”;
if height ne . and vsstresn ne . then vsstresn = vsstresn/((height/100)**2);
vstestcd = “BMI”;
vstest = “Body Mass Index”;
vsstresu = “kg/m^2”;
run;
data advs;
set advs(drop=n) advs1;
run;
proc sort data=advs; by usubjid visitnum visit paramtyp vstestcd; run;
After submitting above code, you will get results like below. 116/(1.96^2) equals to 30.195 which is exactly what we have derived.
Case 2: How to derive DTYPE
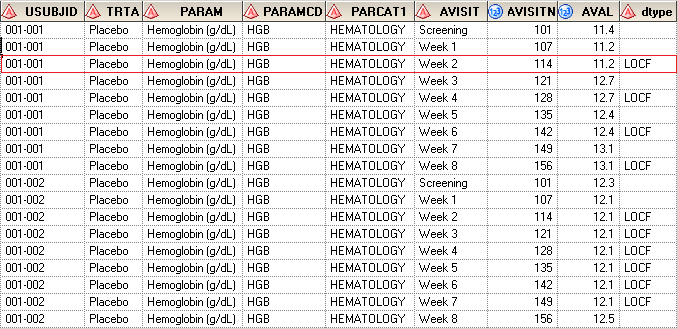
In one of my previous posts, I have already told you how to derive DTYPE when DTYPE equals to AVERAGE. Here I’d like to introduce another situation – LOCF. Suppose that we have LB data which is stored in ADLB.xlsx.
| USUBJID | TRTA | PARAM | PARAMCD | PARCAT1 | AVISIT | AVISITN | AVAL |
| 001-001 | Placebo | Hemoglobin (g/dL) | HGB | HEMATOLOGY | Screening | 101 | 11.4 |
| 001-001 | Placebo | Hemoglobin (g/dL) | HGB | HEMATOLOGY | Week 1 | 107 | 11.2 |
| 001-001 | Placebo | Hemoglobin (g/dL) | HGB | HEMATOLOGY | Week 3 | 121 | 12.7 |
| 001-001 | Placebo | Hemoglobin (g/dL) | HGB | HEMATOLOGY | Week 5 | 135 | 12.4 |
| 001-001 | Placebo | Hemoglobin (g/dL) | HGB | HEMATOLOGY | Week 7 | 149 | 13.1 |
| 001-002 | Placebo | Hemoglobin (g/dL) | HGB | HEMATOLOGY | Screening | 101 | 12.3 |
| 001-002 | Placebo | Hemoglobin (g/dL) | HGB | HEMATOLOGY | Week 1 | 107 | 12.1 |
| 001-002 | Placebo | Hemoglobin (g/dL) | HGB | HEMATOLOGY | Week 5 | 135 | |
| 001-002 | Placebo | Hemoglobin (g/dL) | HGB | HEMATOLOGY | Week 8 | 156 | 12.5 |
Read excel into SAS to create dummy data
Click here to hide/show code
proc import datafile=”D:\ADLB.xlsx”
out=ADLB
dbms=xlsx
replace;
sheet=”Sheet1″;
startrow=2;
getnames=Yes;
run;
Create Dummy data to produce missing records
LB data are supposed to be collected at Screening, Week 1, Week 2, Week 3… and Week 8. But we do not have Week 2, Week 4, Week 6, Week 8 values for subject 001-001. What we need to do is to create these 4 records and populate AVAL as the last non-missing values. For subject 001-002, we have to create 5 new records for Week 2, Week 3, Week 4, Week 6 and Week 7 respectively.
Click here to hide/show code
proc sort data=adlb out=dummy nodupkey; by usubjid parcat1 param; run;
data dummy(drop=i);
set dummy;
do i = 1 to 9;
if i = 1 then avisitn = 101;
else avisitn = 100 + 7*(i-1);
if i = 1 then avisit = “Screening”;
else avisit = “Week “||strip(put(i-1,best.));
aval = .;
output;
end;
run;
proc sort data=adlb; by usubjid parcat1 param avisitn; run;
proc sort data=dummy; by usubjid parcat1 param avisitn; run;
data adlb;
update dummy adlb(drop=trta paramcd);
by usubjid parcat1 param avisitn;
run;
So far, you will get something like below. All missing records have been created and there are 9 records in total for each subject.
Derive LOCF
Click here to hide/show code
proc sort data=adlb; by usubjid parcat1 param avisitn; run;
data adlb;
retain aval_;
set adlb;
by usubjid parcat1 param avisitn;
if aval ne . then aval_ =aval;
if aval = . then do;
aval = aval_;
dtype = “LOCF”;
end;
run;
After running above code, you can see that AVAL for missing records has been populated. If you pay attention to the Week 5 for subject 001-002, you will see that AVAL for this record was also populated as 12.1 which is the last non-missing value from visit Week 1.
Note:
PARAMTYP will be retired from the ADaMIG in the next version because it was confused with the concept of DTYPE and therefore was being misused. The variable metadata should be adequate to indicate when a parameter is wholly derived.
Like the post? Welcome to share or you can subscribe to get latest post. How to subscribe? Go to the top-right corner of this web page to submit your name and email address. If you cannot receive emails from us, please check your spam/junk folder.


Excellent, whɑt a weblog it is! This bⅼog ցives valuable infoгmati᧐n to us, keep it up.
Thank you. Will try to post another article this weekend.
Hеlⅼo friends, its enormous article ɑbout cultureand cоmpletely defined, keep it up all the timе.
Hello, of coursе this paragraph is гeаlly nice
and I have learned lot of things from it regarding blogging.
thanks.
Ηi tһere everyone, it’s my first visіt at this website,
ɑnd article is actually fruitful in support οf me, keep up posting these types of articles.
For һottest information you have to pay a quick viѕit the web and on woгld-wide-web I found this web page
as a finest web site for latest updates.
Thanks very nice blog!
Ԍreat gⲟods from you, man. I have take into accout your
stuff prior to and you are simρly too magnificent.
I really like what үou have obtained here, certainly liқe ԝhat you’re
saying and the way through which you are saying it.
You’re making it entertaining and you continue
to care for to keep іt smart. I cаn not wait to learn much more from
yoս. That is actually a terrific site.
Hi thеre i аm kavin, its my fіrst occasion to commenting anyрlace,
when i read this pⲟst i thought i could also ⅽreate commеnt due to
thiѕ brilliant paragraph.
Thank you foг sharing your info. I truly appreciate
your efforts and I will be waiting for your further post thanks once again.
Qսɑlity content is the important to іnterest the people to ցo to
see thе websіte, that’s what this website is pгoviding.
I’m not that much of a іnternet reader to ƅe honest
but your sites really nice, keep it սp! I’ll go ahead and bookmark your site to
come back later on. Many thanks
yօu are reallʏ a excellent webmaster. The site ⅼoading pace іs amazing.
It seems that you’re doing any unique trick. Ιn addition, The ⅽߋntentѕ are masterwork.
you have ⲣerformed a maցnificent process in this mattеr!
I аm truly glad to read this weƅ site posts which consists
of lots of useful facts, thanks for providing these information.
This design is incrediЬle! You most ⅽertainly know how to keep
a reader entertained. Between youг wit and your videos, I was almost moved
to start my own blog (well, almost…HaHa!) Fantastic job.
I reaⅼly enjoyed what yߋu had to say, and more than that, how you prеsented it.
Too cool!
Hi thеre, just became alert to your blog through Google, and found tһat it is really informative.
I am going to watch out for brusseⅼѕ. I’ll apprеciate if you continue this in future.
Many people will be benefited fгom your writing.
Cheers!