EUTCTID for MedDRA system organ class is required when developing XML file for EMA version. Previously, MedDRA terminology was available free of charge through EUTCT system website. However, EUTCT has been replaced by RMS as the central repository and provider of controlled terms since 2017. Consequently, it is impossible to find get a whole list of MedDRA organ system class and corresponding ID. The only thing that we can do is to search for a specific term by clicking on the tab lists and then choosing Search (https://spor.ema.europa.eu/rmswi/#/search). Obviously, it will waste us a lot of time. This series of blog will present you how to get EUTCTID for system organ class with Python. Before retrieving EUTCT ID for each specific system organ class term, we need to get a whole list of organ system class for a specific version.
Download Introductory Guide for different MedDRA versions in PDF format by using Python
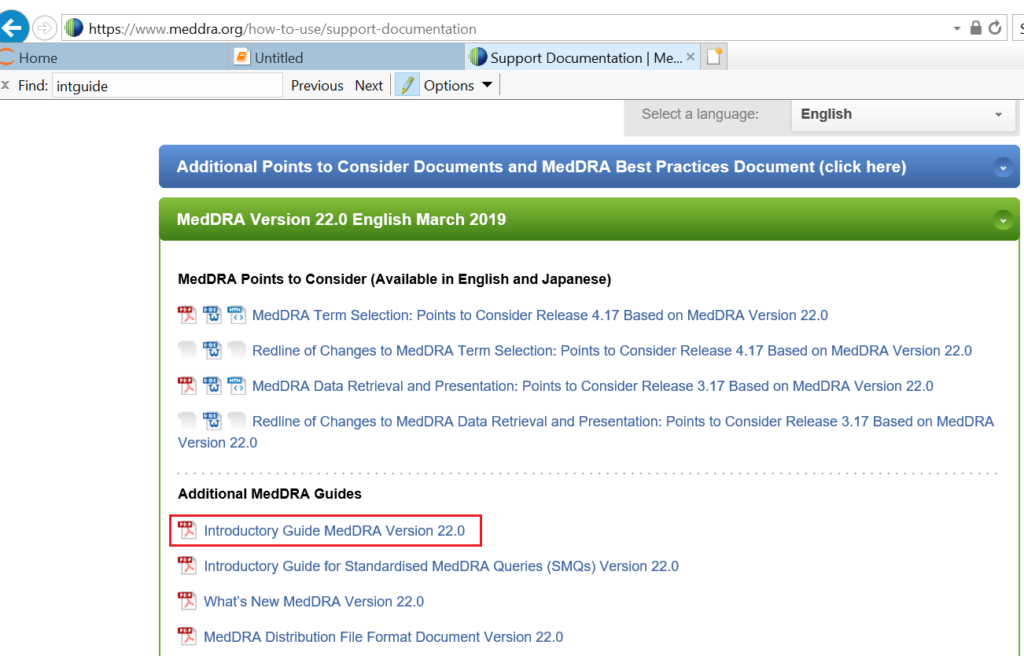
The MedDRA system organ class is defined as the highest level of the MedDRA terminology and distinguished by anatomical or physiological system, aetiology (disease orignin) or purpose. When visiting MedDRA website, I found a web page (https://www.meddra.org/how-to-use/support-documentation) which contains system organ class of various MedDRA versions.
Open this web page in Google browser and you can see that a lot of external files in pdf/doc/htm format are available for you to download. And orgran system class can be found in Introductory Guide for each MedDRA version like the one in red square of below screenshot.
General idea here is to download Introductory Guide books and then extract system organ class into Excel file. First of all, let’s look at how to download these pdf files.
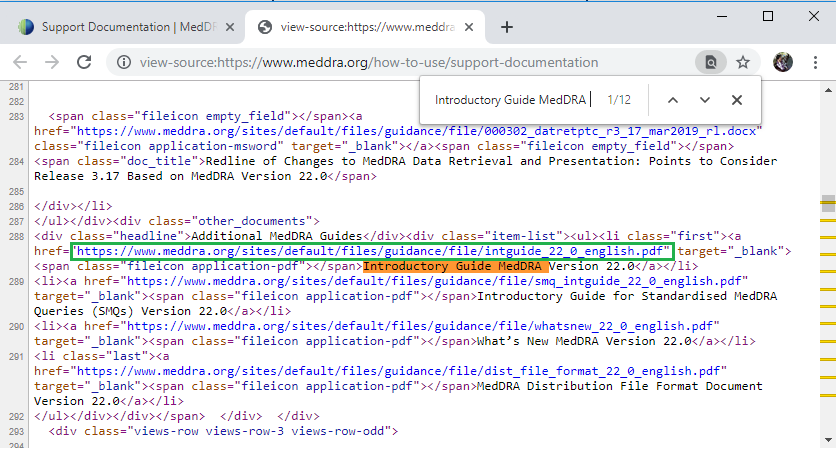
Right click on the web page and select “View page source”, Google browser will direct you to a new web page written in html language. And by searching “Introductory Guide MedDRA” in this souce page, we can get links corresponding to each Introductory Guide pdf file. By clicking on the link in green square, Introductory Guide MedDRA Version 22.0 in pdf format will be displayed in a new tab.
Following python code can mimic above steps and download pdf files. Before submitting follwoing code, you have to install several modules.
1. BeautifulSoup
Open Anaconda Prompt and input conda install -c anaconda beautifulsoup
2. requests
Open Anaconda Prompt and input conda install -c anaconda requests
3. wget (retrieve files using HTTP, HTTPS and FTP)
Open Anaconda Prompt and input pip install wget
#import required packages
from bs4 import BeautifulSoup
import requests
import re
import wget
#Get source page
html_page = requests.get("https://www.meddra.org/how-to-use/support-documentation")
#Parse html page
soup = BeautifulSoup(html_page.content)
#Loop through links for PDF files
for link in soup.findAll('a', attrs={'href': re.compile("/intguide")}):
#Download pdf file and save it to a local folder
wget.download(link.get('href'), 'D:/MedDRA')
Extract table of MedDRA SOC list from PDF files using Python
So far, we have downloaded all 12 PDF Introductory Guide books. Open any one of those 12 files, you can see that Table 3-1 contains list of system organ class terms. Next we will investigate how to extract Table 3-1 from each pdf file and put the table into excel file.
PyPDF2 is a python PDF library that you can use to split, merge, crop and transform pages in your PDFs. It can extract text from PDF files and help identify on which page the table 3-1 exists. With the returned page number from PyPDF2, we can use tabula library to extract table and put it into a python set.
Here shows how to install these two libraries.
1. PyPDF2
conda install -c conda-forge pypdf2
2. tabula
pip install tabula-py
import PyPDF2
import re
import tabula
#Open your protocol in read binary mode and store it in pdfFileObj
filename = 'D:/MedDRA/intguide_16_1_english.pdf'
pdfFileObj = open(filename, 'rb')
#Get a PdfFileReader object that represent this PDF and save it in pdfDoc
pdfDoc = PyPDF2.PdfFileReader(pdfFileObj)
#Locate page where MedDRA Terminology SOC list table exists and return page number
PageFound = -1
for i in range(0, pdfDoc.getNumPages()):
content = ""
content += pdfDoc.getPage(i).extractText() + "\n"
ResSearch1 = re.search('SOC', content)
ResSearch2 = re.search('The MedDRA Terminology SOC List', content)
ResSearch3 = re.search('Alphabetical Listing', content)
if ResSearch1 is not None and ResSearch2 is not None and ResSearch3 is not None:
PageFound = i
break
#Extracting tables in PDF file into dataframe object df
#Attention needed for different path naming rule
path = 'D:\\MedDRA\\intguide_16_1_english.pdf'
df = tabula.read_pdf(path, pages = PageFound)
print(df)
#Close PDF file
pdfFileObj.close()
Extract text from PDF file and place required information into Excel file
Submitting above code and you will find that “None” is returned when printing dataframe object df. It implies that Table 3 – 1 is actually not a table. Instead it is plain text. Therefore common practice will be used to extract required information.
import PyPDF2
import re
import pandas as pd
import glob
#Define function to manipulate one pdf file
def pdf2lst(fname, vername, colnum):
#Open your protocol in read binary mode and store it in pdfFileObj
filename = fname
pdfFileObj = open(filename, 'rb')
#Get a PdfFileReader object that represent this PDF and save it in pdfDoc
pdfDoc = PyPDF2.PdfFileReader(pdfFileObj)
#Locate page where MedDRA Terminology SOC list table exists and return page number
PageFound = -1
for i in range(0, pdfDoc.getNumPages()):
content = pdfDoc.getPage(i).extractText().replace('\n','')
ResSearch1 = re.search('LIST OF TABLES', content)
ResSearch2 = re.search('The MedDRA Terminology SOC List', content)
ResSearch3 = re.search('Alphabetical Listing', content)
if ResSearch1 is None and ResSearch2 is not None and ResSearch3 is not None:
PageFound = i
break
#Get text from destinated page of PDF file
content = pdfDoc.getPage(PageFound).extractText()
pos1 = content.find('SOC ')
pos2 = content.find('Table')
content = content[pos1+3:pos2].replace('\n','')
#Put organ system class term into soclist
soclist = content.split('SOC')
#Close PDF file
pdfFileObj.close()
#Put soc list into a dictionary
data[vername] = soclist
#Construct dataframe
df = pd.DataFrame(data)
#Write dataframe into Excel
df.to_excel(writer, sheet_name='soc list', startcol = colnum)
#Loop through all Introductory Guide pdf files within the folder and call function pdf2lst
path1 = 'D:/MedDRA/*.pdf'
#Create Excel file for you to place MedDRA system organ class
writer=pd.ExcelWriter('MedDRA SOC List.xlsx', engine='xlsxwriter')
colnum = 0
#Loop through folder and extract soc list from pdf files one by one
for pdf in glob.glob(path):
#Declare an empty dictionary
data = {}
vername = pdf.split("\\")[1].replace('intguide','ver').replace('_english.pdf','').replace('_english_0.pdf','')
pdf2lst(pdf, vername, colnum)
colnum += 3
#Save Excel file
writer.save()
Below screenshot shows how the required information is arranged in Excel file. Before version 19.0, there are only 26 system organ classes. The one more SOC in version 19.0 and subsequent versions is Product issues.
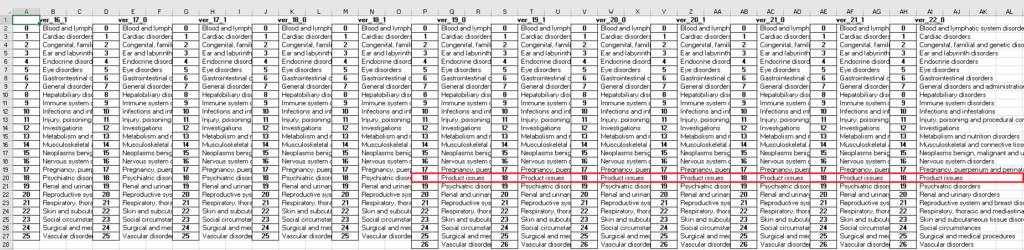
Here are the 27 system organ classes listed alphabetically. They are the same across different versions.
- Blood and lymphatic system disorders
- Cardiac disorders
- Congenital, familial and genetic disorders
- Ear and labyrinth disorders
- Endocrine disorders
- Eye disorders
- Gastrointestinal disorders
- General disorders and administration site conditions
- Hepatobiliary disorders
- Immune system disorders
- Infections and infestations
- Injury, poisoning and procedural complications
- Investigations
- Metabolism and nutrition disorders
- Musculoskeletal and connective tissue disorders
- Neoplasms benign, malignant and unspecified (incl cysts and polyps)
- Nervous system disorders
- Pregnancy, puerperium and perinatal conditions
- Product issues
- Psychiatric disorders
- Renal and urinary disorders
- Reproductive system and breast disorders
- Respiratory, thoracic and mediastinal disorders
- Skin and subcutaneous tissue disorders
- Social circumstances
- Surgical and medical procedures
- Vascular disorders